Search Results
Modis QC Bits
In the course of working through my MODIS LST project and reviewing the steps that Imhoff and Zhang took as well has the data preparations other researchers have taken ( Neteler ) the issue of MODIS Quality control bits came up. Every MODIS HDF file comes with multiple SDS or multiple layers of data. For MOD11A1 there are layers for daytime LST, Night LST, the time of collection, the emissivity from ch 31 and 32, and two layer for Quality Bits. The various layers of MODIS11A1 file is available here. Or if you are running the MODIS package, you can issue the getSds() call and pass it the name of a HDF file.
As Neteler notes the final LST files contain pixels of various quality. Pixels that are obscured by clouds are not produced, of course, but that does not entail that every pixel in the map is of the same quality. The SDS layers for QC — QC_day and QC_Night– contain the QC bits for every lat/lon in the LST map. On the surface this seem pretty straighforward, but deciding the bits can be a bit of a challenge. To do that I considered several source.
2. The Modis groups user guide
4. LP DAAC Tools, specifically the LDope tools provided by guys at MODLAN
I will start with 4, the ldope tools, If you are a hard core developer I think these tools would be a great addition to your bag of tricks. I won’t go into a lot of detail, but the tools allow you to do a host of processing on HDF files using a command line type interface. They provide source code in c and at some point in the future I expect that I would want to either write an R interface to these various tools or take the C code directly and wrap it in R. Since they have the code for reading HDF directly and quickly it might be a source to use to support the direct import of HDF into raster. However, I found nothing to really help with understanding the issue of decoding the QC bits. You can manipulate the QC layer and do a count of the various flags in the data set. Using the following command on all the tiles for the US I was able to poke around and figure out what flags are typically reported
comp_sds_values -sds=QC_Day C:\\Users\\steve\\Documents\\MODIS_ARC\\MODIS\\MOD11A1.005\\2005.07.01\\MOD11A1.A2005182.h08v05.005.2008040085443.hdf
However, I could do the same thing simply by loading a version of the whole US into raster and using the freq() command on raster. Figuring about what values are in the QC file is easy, but we want to understand what they mean. Let’s begin with that list of values. By looking at the frequency count of the US raster and by looking at a large collection of HDf files I found the following values for the QC layer
values <-c(0,2,3,5,17,21,65,69,81,85,129,133,145,149,193)
The question is what do they mean. The easiest one to understand is 0. A zero means that this pixel is highest quality. If we want only the highest quality pixels, then we can use the QC map, turn it into a logical mask and apply it to the LST map such that we only keep pixels that have 0 for their quality bits. There, job done!
Not so fast. We throw away many good pixels that way. To understand the QC bits lets start with the table provided
If you are not clear on how this works every number, every integer in the QC map is an unsigned 8 bit integer. The range of numbers is 0 to 255. The integers represent bit codes which requires you to do some base 2 math. That is not the tricky part. The number 8 for example would be “0001” and 9 would be 1001. If you are unclear about binary representations of integers I suppose google is your friend.
Given the binary representation of the integer value we are then in a position to understand what the quality bits represent, but there is a minor complication which I’ll explain using an example. Let’s take the integer value of 65. In R the way we turn this into a binary representation is by using the call intToBits.
intToBits(65)
[1] 01 00 00 00 00 00 01 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00
[25] 00 00 00 00 00 00 00 00
We only need 8 bits, so we should do the following
> intToBits(65)[1:8]
[1] 01 00 00 00 00 00 01 00
and for more clarity I will turn this into 0 and 1 like so
> as.integer(intToBits(65)[1:8])
[1] 1 0 0 0 0 0 1 0
So the first bit ( or bit 0 ) has a value of 1, and seventh bit ( bit 6 ) has a value of 1. We can check our math 2^6 =64 and 2^0 = 1, so it checks. Don’t forget that the 8 bits are numbered 0 to 7 and each represents a power of 2.
If you try to use this bit representation to understand the QC “state”, you will go horribly wrong!. It’s wrong because HDF files are written in big endian format. If you are not familiar with that, what you need to understand is that the least significant bit is on the right. In the example above the zero bit is on the left, so we need to flip the bits so that bit zero is on the right . Little Endian goes from right to left, 2^0 to 2^7. Big Endian goes left to right 2^7 to 2^0.
Little Endian: 1 0 0 0 0 0 1 0 = 65 2^0 + 2^6
Big Endian 0 1 0 0 0 0 0 1 = 65 2^6 + 2^0
The difference is this:
If we took 1 0 0 0 0 0 1 0 and ran it through the table, the table would indicate that the first two bits 10 would mean the pixel was not generated, and bit 1 in the 6th position would indicate an LST error of <= 3K. Now flip those bits
> f<-as.integer(intToBits(65)[1:8])
> f[8:1]
[1] 0 1 0 0 0 0 0 1
This bit order has the first two bits as 01 which indicates that the pixel is produced, but that other quality bits should be checked. Since bit 6 is 1 that indicates the pixel has an error > 2Kelvin but less than 3K
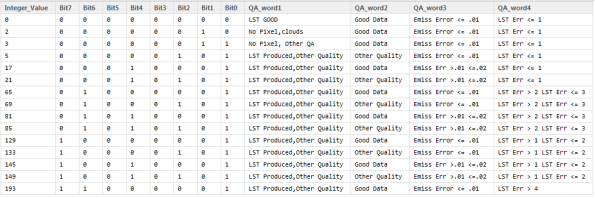
In short, to understand the QC bits we first turn the integer into a bit notation and then we flip the order of the bits and then use the table to figure out the QC status. For MOD11A1 I wrote a quick little program to generate all the possible bits and then add descriptive fields. I would probably do this for every Modis project I worked on especially since I dont work in binary everyday and I made about 100 mistakes trying to do this in my head.
QC_Data <- data.frame(Integer_Value = 0:255,
Bit7 = NA,
Bit6 = NA,
Bit5 = NA,
Bit4 = NA,
Bit3 = NA,
Bit2 = NA,
Bit1 = NA,
Bit0 = NA,
QA_word1 = NA,
QA_word2 = NA,
QA_word3 = NA,
QA_word4 = NA
)
for(i in QC_Data$Integer_Value){
AsInt <- as.integer(intToBits(i)[1:8])
QC_Data[i+1,2:9]<- AsInt[8:1]
}
QC_Data$QA_word1[QC_Data$Bit1 == 0 & QC_Data$Bit0==0] <- “LST GOOD”
QC_Data$QA_word1[QC_Data$Bit1 == 0 & QC_Data$Bit0==1] <- “LST Produced,Other Quality”
QC_Data$QA_word1[QC_Data$Bit1 == 1 & QC_Data$Bit0==0] <- “No Pixel,clouds”
QC_Data$QA_word1[QC_Data$Bit1 == 1 & QC_Data$Bit0==1] <- “No Pixel, Other QA”
QC_Data$QA_word2[QC_Data$Bit3 == 0 & QC_Data$Bit2==0] <- “Good Data”
QC_Data$QA_word2[QC_Data$Bit3 == 0 & QC_Data$Bit2==1] <- “Other Quality”
QC_Data$QA_word2[QC_Data$Bit3 == 1 & QC_Data$Bit2==0] <- “TBD”
QC_Data$QA_word2[QC_Data$Bit3 == 1 & QC_Data$Bit2==1] <- “TBD”
QC_Data$QA_word3[QC_Data$Bit5 == 0 & QC_Data$Bit4==0] <- “Emiss Error <= .01”
QC_Data$QA_word3[QC_Data$Bit5 == 0 & QC_Data$Bit4==1] <- “Emiss Err >.01 <=.02”
QC_Data$QA_word3[QC_Data$Bit5 == 1 & QC_Data$Bit4==0] <- “Emiss Err >.02 <=.04”
QC_Data$QA_word3[QC_Data$Bit5 == 1 & QC_Data$Bit4==1] <- “Emiss Err > .04”
QC_Data$QA_word4[QC_Data$Bit7 == 0 & QC_Data$Bit6==0] <- “LST Err <= 1”
QC_Data$QA_word4[QC_Data$Bit7 == 0 & QC_Data$Bit6==1] <- “LST Err > 2 LST Err <= 3”
QC_Data$QA_word4[QC_Data$Bit7 == 1 & QC_Data$Bit6==0] <- “LST Err > 1 LST Err <= 2”
QC_Data$QA_word4[QC_Data$Bit7 == 1 & QC_Data$Bit6==1] <- “LST Err > 4”
Which looks like this
Next, I will remove those flags that don’t matter. All good pixels, all pixels that don’t get drawn, and those where the TBD bit is set high. What I want to do is select all those flags where the pixel is produced but the pxel quality is not the highest quality
FINAL <- QC_Data[QC_Data$Bit1 == 0 & QC_Data$Bit0 ==1 & QC_Data$Bit3 !=1,]. Below see a part of this table
And then I can select those that occur in my HDF files.
Looking at LST error which matters most to me, the table indicates that I can use pixels that have a QC value of 0,5,17 and 21. I want to only select pixels where LSt error is less than 1 K
Very quickly I’ll show how one uses raster functions to apply the QC bits to LST. using MODIS R and the function getHdf() I’ve downloaded all the tiles for US for every day in July 2005. For July 1st, I’ve used MRT to mosiac and resample all the SDS. I use Nearest neighbor to insure that I dont average quality bits. The mosaic is the projected from a SIN projection to a Geographic projection ( WGS84) and a geotiff output format. Pixel size is set at 30 arc seconds
The SDS are read in using R’s raster
library(raster)
## get every layer of data in the July1 directory
SDS <- list.files(choose.dir(),full.names=T)
## for now I just work with the Day data
DayLst <- raster(SDS[7])
DayQC <- raster(SDS[11])
## The fill value for LST is Zero. That means Zero represents a No Data pixel
## So, we force those pixels to NA
DayLst[DayLst==0]<-NA
NightLst[NightLst==0]<-NA
## Units in LST need to be scaled to be put into Kelvin. The adjustment value is .02. See the docs for these values
## every SDS has a different fill value and a different scaling value.
DayLst <- DayLst * .02
Now, we can plot the two raster
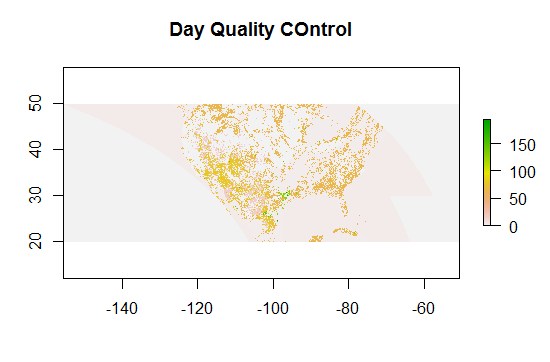
plot(DayLst, main = “July1 2005 Day”)
plot(DayQC, main=”Day Quality COntrol”)
And I note that the area around texas has a nice variety of QC bits, so we can zoom in on that
Next we will use the QC layer and create mask
m <- DayQC
m[m > 21]<-NA
m[ m == 2 | m== 3]<-NA
Good <- mask(DayLst, m)
For this mask I’ve set all those grids with QC bits greater than 21 to NA. I’ve also set QC pixels with a value of 2 and 3 to NA. This step isnt really necessary since QC values of 2 and 3 mean that the pixel doesnt get produce, but I want to draw a map of the mask which will show which bits are NA and which bits are not NA. The mask() function works like so. “m” is laid over DayLst. bits that are NA in “m” will be NA in the destination (“Good”) if a pixel is not NA in “m” and has a value like, 0 or 17 or 25, then the value of the source (DayLst) is written into the destination.
Here are the values in my mask
> freq(m)
value count
[1,] 0 255179
[2,] 5 12
[3,] 17 8539
[4,] 21 3
[5,] NA 189147
So the mask will “block” 189147 pixels in DayLst from being copied from source to destination, while allowing the source pixels that have QC bits of 0,5,17,21 to be written. These QC bits are those that represent an LST error of less than 1K
When this mask is applied the colored pixels will be replaced by the values in the “source”
And to recall, these are all the pixels PRIOR to the application of the QC bits
As a side note, one thing I noticed when doing my SUHI analysis was that the LST data had a large variance over small areas. Even when I normalized for elevation and slope and land class there were pixels that were 10+ Kelvin warmer than the means. Essentially 6 sigma cases. That, of course lead me to have a look at the QC bits. Hopefully, this will improve some of the R^2 I was getting in my test regressions.
Checking the MODIS Install
In the previous step we checked out MODIS install. The key here is making sure that .MODIS_Opts.R has the correct values and is copied the right location. Every time you load MODIS ( library(MODIS)), the current version of .MODIS_Opt.R will be loaded and those defaults will be used for all your processing.
Next, start your R Studio and load the MODIS library. We are going to check out some other commands: checkDeps(); .checkTools() and .getDef(). These functions are “hidden from view” and you don’t usually use them in programming with the package, but since we have the source we can see that they exist, and calling them will give us some more information about the health of our install. To invoke these calls we preceed them with a qualifier MODIS::: Take a look at the console commands below
> library(MODIS)
> checkDeps()
Error: could not find function “checkDeps”
> MODIS:::checkDeps()
Ok all suggested packages are installed!
> MODIS:::.checkTools()
Checking availabillity of MRT:
‘MRT_HOME’ found: c:/Users/steve/documents/MRT
‘MRT_DATA_DIR’ found: c:/Users/steve/documents/MRT/data
MRT enabled, settings are fine!
Checking availabillity of ‘FWTools/OSGeos4W’ (GDAL with HDF4 support for Windows):
OK, GDAL 1.9.2, released 2012/10/08 found!
> MODIS:::.getDef()
After the last command you should see a listing of all your MODIS options settings. For now these are controlled by the file .MODIS_Opts.R. If you want to change the organization of your files there is a command to do that “orgStruc()”. This command allows you to move an entire directory structure to a new location. Before we play with that command, we will need to get some files.
A first attempt at getting files. For this we will use runGDAL(). This command will create the local archive, download hdf files to it and run a gdal process on those file creating processed output. for brevity I’ve shortend some of the output. This is taken straight from the sample code.
runGdal( product=”MOD11A1″, extent=”austria”, begin=”2010001″, end=”2010005″, SDSstring=”101″)
No output ‘pixelSize’ specified, input size used!
No ‘resamplingType’ specified, using default: near
No ‘outProj’ specified, using GEOGRAPHIC
No ‘job’ name specified, generated (date/time based)): C:/Users/steve/Documents/MODIS_ARC/PROCESSED/MOD11A1.005_20121125115753
Getting file from: LPDAAC
trying URL ‘ftp://e4ftl01.cr.usgs.gov/MOLT/MOD11A1.005/2010.01.01/MOD11A1.A2010001.h18v04.005.2010007011052.hdf’
using Synchronous WinInet calls
opened URL
downloaded 1.7 Mb
Downloaded by the first try!
The process will continue to download files and has the wonderful feature of retrying downloads that fail. My internet was acting up a bit so one file took 23 tries. I especially like this feature of the package. When writing simple downloads from ftp I often find that my calls to download.file() fail
Getting file from: LPDAAC
trying URL ‘ftp://e4ftl01.cr.usgs.gov/MOLT/MOD11A1.005/2010.01.02/MOD11A1.A2010002.h18v04.005.2010007015858.hdf’
using Synchronous WinInet calls
opened URL
downloaded 4.1 Mb
Downloaded after 23 retries!
As you watch this command execute you will also see output from the “GDAL” process. That will look like this. if you dont see this output, then your path to GDAL is messed up. ( ha, I messed mine up ) make sure your FWToolspath in MODIS_Opts.R is set to the right location. I have both FWTools and OSGeo4W on my system and I set FWTools to the OSGeo4w version of GDal.
Creating output file that is 891P x 307L.
Processing input file HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~1.HDF:MODIS_Grid_Daily_1km_LST:LST_Day_1km.
Using internal nodata values (eg. 0) for image HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~1.HDF:MODIS_Grid_Daily_1km_LST:LST_Day_1km.
0…10…20…30…40…50…60…70…80…90…100 – done.
Processing input file HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~2.HDF:MODIS_Grid_Daily_1km_LST:LST_Day_1km.
Using internal nodata values (eg. 0) for image HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~2.HDF:MODIS_Grid_Daily_1km_LST:LST_Day_1km.
0…10…20…30…40…50…60…70…80…90…100 – done.
Creating output file that is 891P x 307L.
Processing input file HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~1.HDF:MODIS_Grid_Daily_1km_LST:Day_view_time.
Using internal nodata values (eg. 255) for image HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~1.HDF:MODIS_Grid_Daily_1km_LST:Day_view_time.
0…10…20…30…40…50…60…70…80…90…100 – done.
Processing input file HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~2.HDF:MODIS_Grid_Daily_1km_LST:Day_view_time.
Using internal nodata values (eg. 255) for image HDF4_EOS:EOS_GRID:C:\Users\steve\DOCUME~1\MODIS_~1\MODIS\MOD11A1.005\201001~1.05\MOD11A~2.HDF:MODIS_Grid_Daily_1km_LST:Day_view_time.
0…10…20…30…40…50…60…70…80…90…100 – done.
Before I explain the runGdal() command and all its options, we want to make sure that the process worked. Your local modis archive should be easy to find. Mine is in my R Home directory.
Top Level
And then when we drill down we will find two directories one for HDF files and a PROCESSED directory for the output from GDAL
And when we look at the processed files we will see all the files created
The runGdal() call has thus gone to datapool, selected out the product I wanted ( MOD11A1 or lst ) selected a region of the world which I was able to specify by name “austria”, selected a time period day 1 through day 5, and selected out a couple layers of that file. Remember a MODIS HDF file has multiple Science data layers. These are specified by SDSstring, more on that later
Modis R: Package tutorial
The MODIS package for R
For people who work in GIS in R there has been a bit of challenge in working with data from Modis. I’ll use an example from my own work on UHI to illustrate. A while back I decided that I wanted to look at albedo data from MODIS. If you want to get MODIS data there are basically two approaches. You can use the web services like REVERB which is basically a web interface that allows you to construct queries for a wide variety of data ( or similar services shown here ) or you can download files directly from the datapool ftp.
For my Albedo project I decided to download directly from ftp because I could write a simple program in R to collect the entries in the ftp directories and then download them over the course of a few hours. It’s essentially the same process I use in my study of SUHI for small cities. Getting the data locally is not a big problem, except for the storage issue. You can imagine what it would take to download 90 days of LST data for the US. with 10-14 tiles of data required that ends up being a nice chunk of data.
The next challenge is getting MODIS data out of its native format into a format that is friend for R GIS packages. In the case of Albedo that data was stored in a format known as “HDF” or HDF-EOS. I work with the raster package and raster ( along with rgdal) have no way of reading HDF directly. This meant I had to find a way to transform HDF into a more common format like “geoTiff”. There was also the issue of tiling multiple hdf files into a single raster. Unlike other datasets that might give you a monolithic file of the whole world, the MODIS collections are distributed as tiles using their tiling system.
To solve this problem there are a couple tools: MRT or the modis reprojection tool. or GDAL, the Geospatial Data abstraction Library. For Albedo I decided to use GDAL and I used the distribution of GDAL provided by FWTools. With a few hundred albedo files I had to write a script to load every Hdf file and transform it to a ‘geotiff’ file. That worked fine, but now I had a set of R code for doing my statistics and a “bat” file for controlling Gdal. With my SUHI project I decided to use MRT. In that case I used the GUI they provide to select all the files, mosaic them, crop them, resample and reproject. I quickly learned that duplicating the same steps with a GUI is not always easy. Further, if I am trying to create work that is reproduceable, it makes it hard. Plus I could not see using that tool to create 365 maps for every day in the year.
What’s needed is an R package that allows us to write R code and achieve the same results as we get with GDAL or MRT. We want to write R code that controls these external applications and tools. That’s what MODIS R does.
The team: The MODIS R team consists of Matteo Mattiuzzi, Jan Verbesselt, Forrest Stevens, Tomislav Hengl, Anja Klisch, Bradley Evans and Agustin Lobo. Some of the seminal work was done by Tomislav at his wonderful page
My Tutorial. I had some discussions with Matteo who is the package maintainer and suggested that I might do a tutorial to help people get started with the package. At this stage the package is stable, so I decided to start a tutorial. The way I work is pretty simple. As I try new things I write them up. For now the tutorial starts with the basics of R, R studio and preparing your system for MODIS R. That includes downloading a the tools and testing them out and finally downloading and building a WINDOWS version of MODIS R.
Then I will start to go through the API and test and explain all the calls in more depth than the manual and write some sample code.
A note. If you have questions about what I am doing go ahead and leave comments. I can collect up questions and those I cant answer I can pass on to the maintainers.
Configuring MODIS R
In The last step you should have built the MODIS package and made a zip file. If you have trouble doing that, you can just download my zip file from the drop box and let me know what trouble you had.
With the MODIS package built, you now have to install it. Within R Studio, click on Packages in the window
click on “Install Packages” and a pop up dialogue will appear. From top selector choose install from “zip”. Then browse to the folder that has your Modis zip package. Install the package and load the library library(MODIS)
The first thing you will be asked to do is to modify a file. .MODIS_Opts.R and you will be directed where to save it. This file contains your default parameters. There is one parameter that you should check. GDALpath.
Here is what my file looks like: I have BOLDED the one line that you need to add setting the GDALpath before saving the file. Make that change and save the file to the location provided. For me it was in Users/steve/Documents which is my R Home.
Below this I will explain what is going on
# This file contains default values for the R package ‘MODIS’.
# version 0
#########################
# This file uses codified ‘placeholders’ for the creation of the local archive structure, or to define the path on an remote location. This ‘placeholders’ are than substituted from internal functions using the name of a EOS-file. Use ‘placeholders’ only for ‘arcStructure’!!! Don’t substitute the ‘placeholdes’ with a specific value.
# Placeholders are:
# PLATFORM = “Terra”,”Aqua”, “Combined”, “SRTM”, or “ENVISAT”
# PF1 = “MOLT”,”MOLA” or “MOTA”
# PF2 = “MOD”,MYD” or “MCD”
# YYYY = year (i.e. ‘2009’)
# DDD = doy of the year (i.e. ‘003’)
# DATE = YYYY.MM.DD (i.e. ‘2009.01.03’)
# SENSOR = the sensor name (i.e. ‘MODIS’)
# CCC = collection (if applicable) as 3 digits character (i.e. ‘005’)
# C = collection (if applicable) as numeric value (i.e. 5)
# PRODUCT = the product name (i.e. ‘MOD13Q1’)
# TIME = HHMM (MODIS Swath data style don’t use it for now)
# “/” = new subdirectory (i.e. PRODUCT/DATE)
# “.” = internal separator (i.e. PRODUCT.CCC)
# Example: The file ‘MOD13Q1.A2009017.h18v04.005.2009036150230.hdf’, the localArcPath<- ‘~/MODIS_ARC’ and arcStructure <-‘/SENSOR/PRODUCT.CCC/DATE’ on a UNIX system will generate the following structure: ‘/home/YOURUSER/MODIS_ARC/MODIS/MOD13Q1.005/2009.01.17/MOD13Q1.A2009017.h18v04.005.2009036150230.hdf’
# Once you have modyfied values in sec 1. use the function ‘orgStruc()’ to re-organise your archive
#########################
# 1.) Path and archive structure defaults:
# set path. All data will be stored below this directory. If it doesn’t exist it is created.
localArcPath <- ‘~/MODIS_ARC’ # Don’t forget to call the function ‘orgStruc()’ after changing here!!
# set path, default output location for GDAL, FWT, SSOAP, MRT processing results. If it doesn’t exist it is created.
outDirPath <- ‘~/MODIS_ARC/PROCESSED’
# define, local archive structure. USE ‘placeholdes’!!
arcStructure <- ‘/SENSOR/PRODUCT.CCC/DATE’ # Don’t forget to call the function ‘orgStruc()’ after changing here!!
#########################
# 2.) Processing defaults:
resamplingType <- ‘NN’
outProj <- ‘GEOGRAPHIC’
#########################
# Windows specific (run: “MODIS:::.checkTools()” for inputs)
# Example:
FWToolsPath <- “C:/OSGeo4W/bin”
#########################
# PLEASE DON’T MODIFY BELOW HERE, NOT IMPLEMENTED YET.
# Example ftpstring
# 3.) If you have a personal MODIS datapool within your LAN and you face problems in the creation of an additional ‘ftpstring’ please contact one of us.
# Use ‘placeholders’ only in ‘variablepath’
# Additional ‘placeholders’:
# DATE1DATE2 period start/end (MERIS specific)
# REGION (MERIS specific) MERIS date is stores in regions
# ftpstring0 <- list(name=’sitename’,SENSOR=’sensorname’, basepath=’/base/path’,variablepath=’/variable/path/’,content=c(‘what data is awailable? ‘images’, ‘metadata’!
ftpstring1 <- list(name = “LPDAAC”, SENSOR = “MODIS”, basepath = “ftp://e4ftl01.cr.usgs.gov”, variablepath = “/PF1/PRODUCT.CCC/DATE/”, content = c(“images”, “metadata”))
ftpstring2 <- list(name = “LAADS”, SENSOR = “MODIS”, basepath = “ftp://ladsftp.nascom.nasa.gov/allData”, variablepath = “/C/PRODUCT/YYYY/DDD/”, content = “images”)
ftpstring3 <- list(name = “Culture-MERIS”, SENSOR = “MERIS”, basepath = “ftp://culturemeris:culturemeris@ionia2.esrin.esa.int”, variablepath = “/DATE1DATE2/REGION/”, content = “images”)
ftpstring4 <- list(name = “JRC.it”, SENSOR = “C-Band-RADAR”, basepath = “ftp://xftp.jrc.it/pub/srtmV4/tiff”, variablepath = NULL, content = “images”)
ftpstring5 <- list(name = “Telescience”, SENSOR = “C-Band-RADAR”, basepath = “http://hypersphere.telascience.org/elevation/cgiar_srtm_v4/tiff/zip”, variablepath = NULL, content = “images”)
ftpstring6 <- list(name = “CGIAR”, SENSOR = “C-Band-RADAR”, basepath = “http://srtm.csi.cgiar.org/SRT-ZIP/SRTM_V41/SRTM_Data_GeoTiff”, variablepath = NULL, content = “images”)
So, what is going on here. This file will be used to make your directory structure for storing your LOCAL MODIS FILES, and the OUTPUTS of any processing you do with MRT or GDAL. Having a controlled local archive of MODIS allows you to control your storage and to insure that you dont end up with duplicate files or files thrown all over the place. Also, the functions we use to interact with GDAL and MRT will know where to write results. For Now, lets not muck about with changing where the files get put.
Add the line for GDALpath and make sure it points to the location where your gdal installation is.
Then save the file to the location indicated. Then close your R session and restart.
More Fun With Modis
I’ve started a tutorial on using the MODIS package in R the first few steps are here. While I wait on a release of the package I thought I would play around a bit with the MRT tool and see how it worked. Let’s recall where we are going. I have an inventory of around 150 small cities and we are going to take a look at SUHI in those locations. I have the DEM data, The Modis Land Class data, Olson Biomes, NLCD 30 meter land class and impervious surface. The last few bits I will need are MODIS LST and perhaps MODIS albedo ( If I get motivated) and EVI. We also have population data from a couple sources and housing data.
The first step to getting MODIS LST data is deciding what Tiles I need and then selecting dates. I had hoped that I might get away with using just a few tiles for the US, but that doesnt look like its going to happen. To figure out which Tiles I need there is a function in the MODIS package called getTile. But before I figured that out I tried to do it on my own. With some help from the geo sig list I have the following to figure out the tiles.
First I get the bounding coordinates for every tile
library(geosphere)
library(sp)
gring <-“http://landweb.nascom.nasa.gov/developers/sn_tiles/sn_gring_10deg.txt”
GRING <- read.table(gring, skip = 7, stringsAsFactors =FALSE, nrows=648,
na.strings=c(“-999.0000″,”-99.0000″))
colnames(GRING)<-c(“v”,”h”,”ll_lon”,”ll_lat”,
“ul_lon”,”ul_lat”,”ur_lon”,”ur_lat”,
“lr_lon”,”lr_lat”)
GRING <- GRING[!is.na(GRING[,3]),]
DF <- GRING[GRING$h < 15 & GRING$h > 6 & GRING$v < 7 & GRING$v > 3,]
The table is downloaded and then I remove those rows that are NA. Next, I select those parts of the table that relate to CONUS between h(horizontal) 15 and 6 and vertical 7 and 3.
Next, I use Robert Hijmans code for generating a spatialpolygondataframe.
m <- as.matrix(DF)
z <- apply(m, 1, function(x) Polygon(matrix(x[c(3:10, 3:4)], ncol=2, byrow=T)))
n <- names(z)
y <- sapply(1:length(n), function(i) Polygons(z[i], n[i]))
sp <- SpatialPolygons(y, proj4string=CRS(‘+proj=longlat’))
spdf <- SpatialPolygonsDataFrame(sp, DF)
then we make some corrections for being on a sphere and we read in the lat/lon of all 150 sites
spdfc <- makePoly(spdf)
Inv <- read.table(“SiteInventory.inv”,stringsAsFactors = FALSE)
The next bit of clumsy code creates a spatial points data frame that has 4 points for every site. I’m going to look +- .5degrees from the site center.
myExtent <- data.frame(lon=rep(NA,4*nrow(Inv)),lat=rep(NA,4*nrow(Inv)))
for( site in 1:nrow(Inv)){
dex <- ((site-1)*4)+1
myExtent$lon[dex]<- Inv$Lon[site] +.5
myExtent$lat[dex]<- Inv$Lat[site] -.5
myExtent$lon[dex+1]<- Inv$Lon[site] +.5
myExtent$lat[dex+1]<- Inv$Lat[site] +.5
myExtent$lon[dex+2]<- Inv$Lon[site] -.5
myExtent$lat[dex+2]<- Inv$Lat[site] -.5
myExtent$lon[dex+3]<- Inv$Lon[site] -.5
myExtent$lat[dex+3]<- Inv$Lat[site] +.5
}
Finally we will intersect the points with the SpatialPolygonsDataframe and get out the tiles
pts <- SpatialPoints(myExtent)
proj4string(pts)<- proj4string(spdf)
Tiles <- over(pts,spdfc)[1:2]
Tiles <- unique(Tiles)
nrow( Tiles)
print(Tiles)
Depending on the increment I used (.5 versus .2) I basically had to collect 10-12 tiles to cover my bases for the next step.
With that list of tiles I downloaded the tiles I needed for July4th 2006. In the future with the MODIS package this will happen just by writing code, but for now I did it by hand. With those 12 tiles downloaded I then fired up MRT to mosaic , resample, and reproject the tiles into a monolithic GeoTiff for daytime LST for the whole Conus.
Reading that it we get the following
And then we can zoom in on the midwest and then go down chicago
On the left at lon -87.9 lat 41.65 is the Paw Paw Woods Nature Preserve. Pretty Cool
Installing MODIS for R
If you have installed MRT and OsGeow4, and reviewed the Modis data on datapool, then you are ready for the next step: Installing MODIS package. At the current writing the package is not posted to CRAN, that will happen in due course. If you want to get started, then, you’ll have to install it from source using bits available at R Forge.
Before I show you how to do that, let me pull up the description file from the package
Package: MODIS
Version: 0.6-22
Date: 16-November-2012
Title: MODIS download and processing package. Processing
functionalities for (multi-temporal) MODIS grid data.
Depends: R (>= 2.10), raster, bitops
Suggests: RCurl, rgeos, XMLSchema, rgdal, mapdata, maptools, plotrix,
SSOAP, XML, snow, ptw
Author: Matteo Mattiuzzi, Jan Verbesselt, Forrest Stevens, Tomislav Hengl, Anja Klisch, Bradley Evans and Agustin Lobo
Maintainer: Matteo Mattiuzzi <matteo.mattiuzzi@boku.ac.at>
Description: MODIS download and processing Package. Downloading, re-projecting, re-sampling, mosaicking, format conversion, SDS-extraction, bit-enconding and filtering/smoothing capabilities. Download data from FTP (local processing) or using SSOAP (online processing) via the MODIS web-service. Pre-processing data with MRT, GDAL, FWT, SSOAP. Starting from version 0.3-14 also SRTMv41 can be downloadad. Manual: https://www.dropbox.com/sh/18t0rgcm6bga7xt/-4k_Xwojxr/MODIS
URL: http://r-forge.r-project.org/projects/modis/
License: GPL (>=3)
LazyLoad: yes
Repository: R-Forge
Repository/R-Forge/Project: modis
Repository/R-Forge/Revision: 341
Repository/R-Forge/DateTimeStamp: 2012-11-16 09:16:33
Date/Publication: 2012-11-16 09:16:33
Packaged: 2012-11-16 11:17:11 UTC; rforge
The package description lets us know what other packages we are going to have to install first to make the MODIS R package work. Using R Studio you need to go get the following packages: raster, bitops,RCurl,rgeos,XMLSchema, rgdal,mapdata,maptools,plotrix,SSOAP,XML,snow and ptw.
The only packages that might be a bit tricky to install are RCurl, and SSOAP. As I recall RCurl should work right out of the box on a windows system. It makes calls to curlib and your windows system should have that installed. SSOAP is a different matter.
In my current testing I havent been able to get any of the calls that rely on SSOAP to work, so I’ll be making bug reports. Still there are plenty of functions that dont need SSOAP. You just need it installed.
SSOAP resides here. This package is an “S” implementation of SOAP. To find out about SOAP.. wiki. I think of SOAP as just another way to get data from a web service. Instead of downloading from an ftp site, some services are set up to respond to SOAP requests or queries. Also, don’t be confused by the fact that SSOAP is a “S’ package. R is just an implementation of S.
When you download the package you will note that it is a source distribution, NOT a typical windows binary package which comes in a zip. That means we will have to build the package locally.
If you don’t know how to build a package on R then see my package tutorial. Specifically, we are going to be doing a windows build.
After you download the SSOAP package to a directory, you will want to “untar” or decompress the tar.gz file. If you don’t have winzip installed you can do this from within R using “untar” make sure to set the compressed variable to TRUE and it will default to unzipping a “gz” file.
With the file unzipped, you now use your windows console. Change directories until you are in the SSOAP directory and then use the command:
R CMD INSTALL –build SOAP_.09-0.tar.gz
This will create the package you need. Then using R Studio, install packages from a local zip. select SSOAP and you are good to go. If this is unclear, let me know and I will expand this section.
With all your packages installed you are ready to download the source code for MODIS. You will go through the same process as with SSOAP. get the source bits and use R CMD INSTALL. If you dont understand how to build packages, please read the tutorial.
The last step here will be getting MODIS bits. The source for the package is located Here
A few agreements however. Since the package has not hit CRAN yet, if you have any issues with the windows version you should probably contact me before you contact Matteo. He is pretty busy working on finalizing things and I don’t think we should interrupt him with too many questions. I’ll be going through the code and functions and doing windows testing as I write this tutorial, so if your are just learning R or struggling with this pre release code, it will probably help to work with me before we bug the developers. Of course you can ignore that and write to him directly.
Download the “tar.gz” file to a directory and then “unzip”. You should have all the source for the package. Next take the same steps as you did with SSOAP. Open a console window. > R CMD –build MODIS_0.6-22.tar.gz That should work to create a ‘zip’ binary. Then open R Studio and install packages from a local zip
Then go to the next step
MODIS Reprojection Tool
Download MRT from here. I just downloaded the zip file to my Desktop. You will have to create a REVERB account or ECHO account to get the tool. Before you start the unzip of the MRT you should take a few steps to prepare yourself. I struggled a lot with this install because I was not prepared, so lets take some time to prepare. This is not a simple unzip the file and click install. The install I do here was for Windows 7, but the installer gives you these choices:
Which version of Windows are you running?
1. Windows XP or later
2. Windows 2000
3. Windows NT
Unless you are in fact running Win2000 or NT then #1 will be the choice you want to make. If you are running Windows2000 then the install is going to make a change to autoexec.bat. If you are running NT, XP or anything later than XP ( Vista, 7 etc) Then there is no autoexec.bat to change and the installer will be modifying other files to do the install.
Other things you should do as a preliminary. Decide where you want to install MRT because the install will unzip files to this directory. And most importantly write down your path to java. So I would start by updating java from www.java.com. After you update java you need to copy down the path to java. You must do this exactly! You need to find the path to java.exe. On my system it is located here.
“c:\Program Files (x86)\Java\jre7\bin\java.exe”
Once you have that, then you should unzip the file you downloaded. That download will contain the following files after unzipping: mrt_install.bat, MRT_WIN.zip, unzip, and reg_set. If you like open up the *bat file in notepad. If you double click on it it will just execute. I suggest reviewing the bat file before running it.
If you double click on it you will see the following screen
Then just follow the instructions. You will be asked to specify a directory to install the files. The MRT_WIN.zip file will be unzipped to that directory. You will also be asked for your java path.
After you finish the install you can also create a shortcut for your desktop. Look in the “bin” folder of MRT. You will find a file named modis_tool.bat. Right click and create a shortcut. The bin directory also has an icon for the shortcut. After creating a shortcut for this file, you can copy it to your desktop.
Click on the shortcut and you will see the following. If you dont create a shortcut, just start the program by doubling clicking on Modis_Tool.bat in the “bin” directory.
Here is the main screen
Click on Open Input File and select the HDF file that you downloaded previously
After loading the file we will reproject it to A Geotif using geographic projection.
Note I have ALL the selected bands in this step. Each band will be resampled and reprojected. I selected an output file name. Resampling I have selected nearest neighbor. Note the QC files are all bit data, so nearest neighbor is the right choice. I’ve selected a geographic pr0jection and I edited the parameters (Edit Projection Parameters ) to give the output WGS84 datum. If you click run the input file will be unpacked into the various bands and each band will be resampled, projected and saved as a geotif.
To show you what the Geotif looks like, I’ll pull it up in R.
library(raster)
library(rgdal)
day <- “test.LST_Day_1km.tif”
LST <- raster(day)
LST <- LST * .02
LST[LST==0]<-NA
Note that I scaled the values in the file by .02. SCale factors are located in the documentation on the file. Then I set 0 to NA. After plotting we see this which is Land Surface Temperature in Kelvin
and then I can crop the data and look at a smaller area. The pixels here are roughly 1km ( 926 meters ) On the bottom we have LST during the day and on the top we have the landcover classes from my other project. The red detail in the middle is a small city.
Getting MODIS Data and Tools
By the end of this tutorial the following steps will all be executed in R using the MODIS package. To gain a little understand and test the tools we download, however, the first thing we are going to do is a manual download of a MODIS file and some MODIS tools and then we will test out those tools to make sure they work as expected.
The files that we will download can be access through the Data Pool which is located here Data Pool Home. At the data pool home you should aquaint yourself with the MODIS product table which is here. Product Table. For this exercise I will be downloading a MODIS product known as MOD11 A2. That is a 1km resolution 8 day average of Land Surface Temperature. MODIS LST 8 day average. That file is located on the Terra FTP site MODIS TERRA FTP I’m going to select the version 5 dataset MOD11A2 Version 5 . Specifically a file from the 8 days starting 4th of July 2006. 2006 4th of July.
To get the file I want I merely use the kmz file provided here Modis Tiles in Google Earth whcih tells me the Tile specification. I want a file that covers tile h10v04. If you don’t have google earth you can also use this image

I will download two files. The *hdf file and the xml file.
Before doing that I can browse a jpg of the file here
Download the following two files to a directory we will call Test.
Next up we will get some tools to help us with these files. You will need to register to get the modis reprojection tool
Download that tool. And read the next page before unzipping it
Beginners Guide: Using MODIS in R
This tutorial is going to assume that you are a beginner in R and Windows and working with MODIS. Obviously, if you already know R and Windows pretty well you can skip a bunch of steps. However, it will be good for you to walk through all the steps as I may do some things in the beginning that influence downstream work. As an overview here is what we are going to do. First we are going to update our R tools and make sure we have the fresh bits. Then we are going to spend some time getting familiar with the bits of the windows system that matter. Basically the console. If you don’t know how to build a package in R under windows, go walk through my package building tutorial.
Then we are going to download some MODIS data by hand and get some tools and utilities for working with MODIS data. If you just have a small job sometime it may be better to work “by hand”. At the end we will start using the MODIS package instead of doing things by hand. In very simple terms the MODIS package allows you to find and download MODIS data using “R” commands. More importantly, it allows you to control the tools, MTR and GDAL, from within R. So, first we do some things by hand downloading a file, using the tools by hand, and then we will use R programming to do the same thing. Trust me, when you have hundreds of MODIS files to download reproject and crop, working by hand is a PITA.
First things first however. I want to thank the MODIS R team for all the hard work they have done.
Matteo Mattiuzzi, Jan Verbesselt, Forrest Stevens, Tomislav Hengl, Anja Klisch, Bradley Evans and Agustin Lobo. Conrats on a great package
UPDATE R
While it’s not necessary to start your process of building a package by updating your version of R to the current version, I started my process that way. Ordinarily I like to stay current with R and with all my tools. For this example I will be using version 2.15.1 The exact url for version 2.15.1 is
http://cran.r-project.org/bin/windows/base/R-2.15.1-win.exe
Its best to use this and actually see what version we are on
http://cran.r-project.org/bin/windows/base/
GET RSTUDIO (optional)
In addition to using R I also use RStudio which is a great cross platform IDE. I recommend it use. After updating my R I updated my RStudio from the following URL
I’m using version .96.331
When you have finished that it’s time to do a little review of your Windows System. For some of the tools we will need updating the PATH variables in windows may be required. If you never worked back in DOS and the early days of windows you may not be familiar with the PATH variable.
In order to build a package in R there are a couple things you need to understand about your Windows System. These are fairly trivial but if your coming from a different platform (like MAC) then you might not know where these things are. I will take some time to take you to a couple places on your Windows system. There are two bits that we are going to get familiar with: The console application, and the PATH variable.
First , we are going to locate the console application. You find this application by going to your start button in the lower left hand corner of your desktop
COMMAND PROMPT
Open the start button select “All Programs” –>”Accessories” –>”Command Prompt”. The old reliable “DOS” Prompt.
PATH
The bit of the windows we may have to track down is the PATH Variable. Again, this is an environmental variable that the windows system uses to locate programs and insure that commands get interpreted correctly. To locate your PATH variable Start by clicking on the Start button. At the start panel select the control panel.
Next Select system and security
Next select System
Next on your left select Advanced system settings
At the bottom select Environmental Variables
In the lower Panel select the “Path Variable” and select Edit
That is how you will access your path variable when it comes time to edit it. At this stage I would copy the PATH variable and paste it to a simple txt document. Just in case we screw anything up and we need to recover. What we will be doing is adding various elements to that string and restarting the computer. So, make a copy now before we start. If you have none completed my tutorial on building windows packages I’ll suggest you do that now
The next step is to locate and download some sample MODIS data. later we will do this step in an automated fashion from inside R, but to gain some understanding of the external tools we will be calling from within R we will start by doing a few things manually.
Installing OSGeo4w
The preferred source for gdal on windows is OSGeo4w. FWTools is no longer being kept current with updates to gdal so I suggest that you install OSGeo4w. I believe that FWTools is built on gdal 1.7 and OSGeo4w ( at this time ) is up to gdal 1.9.2. So, we use OSGeo4w to stay current with gdal.
First, familarize yourself with gdal. The web page is here. Gdal is a open source data translation tool. In order to work with MODIS data you need a mechanism for getting it out of “hdf” or hdf eos” format into a format that can be used by R’s GIS packages. So, for example, if you want to use “raster” you need to transform the “hdf” formated data into something readable by raster like “geotiff”. gdal is the tool used to do that.
Gdal comes with various utilities, listed here: so, utilities like gdal_transform and gdal_warp can be used to transform and reproject data prior to importing it into R. Of course, some of these tools are available within R ( using rgdal, sp, raster), but for MODIS R, and big jobs, you probably want to use the gdal tool as an external program. Basically, as long as you are reading in a “hdf” file using gdal, you should apply whatever other processing you want to do at that point. More specifically, “rgdal” does not have the ability to read “hdf”, so you have to use the external version of gdal to do this. I hope that makes sense. MODIS comes in “hdf” format. To read that format and output geotiff you can use MRT or Gdal. MRT will allow you to mosaic tiles, resample, reproject, and output. That output is then readable by “raster”. Gdal has more capabilities than MRT and also outputs geotiff. So, you have two paths for getting data into R friendly formats: MRT and gdal. Both MRT and gdal can be used manually ( as shown in the MRT example and the FWTools example ) OR you can write scripts to execute bigger jobs. The purpose of MODIS R, is to allow you to write these “scripts” or “jobs” in an R environment. So in the end we will use MODIS R to make calls, like runGdal(). those calls will invoke the external tool. To recap you can:
1. Use MRT or gdal manually to transform “hdf” into R friendly formats.
2. you can write “scripts” to control MRT or gdal.
3. You can use MODIS R to make “calls” that execute the external tools.
Now, go here to get OSGeo4W.
OSGeo4W is basically an installer that installs a variety of open source GIS tools like GRASS, QGIS and also gdal. You get the installer at this location
Download the installer. Select express install. Unless you are interested in GRASS or QGis or the other tools, just deselect them and only install GDAL/OGR.
The install will happen at the root, so on my machince I end up with SGeo4w installed at C://Osgeo4w. It will be important to remember where you installed it. MOST IMPORTANT is the fact that the gdal libraries are in the “bin” directory, so C://OSGeo4w/bin will be your likely path to gdal. The “pathGDAL” variable is one you are going to have to set when you use MODIS R, because MODIS R is going to make “calls” to gdal.
If you are a beginner in R you are probably not familar with the system call: system(). MODIS R, uses system() to make calls that the windows OS will then pass to gdal or MRT. That is how we execute gdal and MRT from within R. We make system() calls. For those calls to work either your windows path has to “know” where the tool is ( MRT works this way, since it sets a HOME_DIR) or in the case of gdal we tell the system call where the tool is located.
After you have OSGeo4W installed, please check that you know where it installed and have a look inside the “bin” directory for gdal.
Mine looks like this