A gWidgets GUI for climate data
If you haven’t worked with the gWidgets package it’s worth some time exploring it which is what I’ve been doing for a little paleo project I’ve been working on. After struggling with the few demos and tutorials I could find I went ahead and bought the book: Programming Graphical User Interfaces in R. Luckily the book was a little better than the tutorials which just scratch the surface. GUI programming, in my mind, is unlike other programming and I had many false starts, primarily struggling with lexical scoping issues and getting widgets to update based on changes made to other widgets and the underlying data. The only reliable method I could find was <<- Yikes. I’m sure over time I’d figure away around making these type of “globals” it ended up looking pretty but being ugly under the hood.
Lets Start with the Top Window
Pretty basic but it took a while to get this to work. Part of the problem was mixing the window on the left a ggraphics() widget with the panel on the right and having it operate correctly with resizing. I tried ggroups(), gframe(), combinations of those two and in all cases the windows would not draw properly upon resizing. Text got left all over the place. That problem was solved with a gpanedgroup(). So the left pane is a gframe() that contains a gcheckboxgroup() and then a ggrpahics() widget and the right group is also a gframe containing other widgets. The purpose of the GI is pretty simple: take a huge climate data file and station inventory and “crop” it with respect to a climate proxy file, in this case pollen data. The idea is to create some custom climate files without requiring programming. The process starts by loading a climate file ( from Berkeley Earth datasets) which uses my BerkeleyEarth Package. Then you load a pollen database and plot the two:
The glabel(0 widgets on the right show you how many stations are loaded and the min and max time. They also show the lat /lon which defaults to the whole world. Next we select our region of interest by using the graphics crosshairs. Selecting that “extent” will automatically crop the climate data to the extent selected:
We see the station count adjust and the lat/lon adjust. And the times at which we have temperatures is displayed. Then hit update plot:
And the plot updates. Here I’ve selected an even smaller region. Unfortunately if you make a region too small, the ONLY way to undo that mistake is to “reset data” which brings you back to square one with all the data present. The last step is to adjust the time window: Here I want to build a dataset from 1961 to 1990. So I move the gsliders() and hit update plot.
The last step I have to write is the “save file”. I also had plans to add more plots, like stations versus time, but at one point I hit a nasty gWidgets error about hitting the limits on the stack. Probably my programming. For now, this will do. For future projects I think I’m going downstream to do toolkit programming. The layout restrictions in gWidgets did cramp my style and I really haven’t mastered the whole “signaling” handler, updating widgets.. perhaps I should use an observer model.. more reading.
I usually post code. In this case its so ugly that it would probably teach you bad things. When I get better at this I’ll do some tutorials.
Updates to Old code
I notice from time to time that people download code from the drop box.
The drop box lags CRAN. I’ve upload the newest versions to the drop box.
In the future I will post new releases to the drop box before I submit to CRAN.
Nick Stokes Distance code, now with Big Memory
In my last post I was struggling with getting a big memory version of the distance matrix to work fast. Nick and other readers had some suggestions and after puttering around with Nicks code I’ve adapted it to big memory and not impacted the run time very much. For comparison writing a 10K by 10K matrix was taking minutes and I projected about 24 hours for berkeley data. Nick came to the rescue with two really cool ideas. The first is a different method for calculating distance on the sphere. Here he expanded on my idea of pre computing sin() and cos() for latitudes and longitudes and he developed an approach that allows us to compute these values once and then solve a distance equation using super fast matrix maths. On top of that he came up with a “block” approach. At first this looks like it will be slower because we calculate the entire matrix as oppsoed to just the upper half. In his approach Nick wrote out files for every block. In my approach I write blocks to a filebacked big matrix. Thats a bit slower but I get one file that I can then random access. Here’s the code
createDistanceMatrix <- function(Inventory, blockSize = 20, Directory= getwd(), filename
= “distance.bin”){
# based on code by Nick Stokes
require(bigmemory)
options(bigmemory.allow.dimnames=TRUE)
if(!grepl(“\\.bin”,filename))stop(“please use a bin extension”)
columnsPresent <- intersect(colnames(Inventory),c(“Id”,”Lon”,”Lat”))
if(length(columnsPresent) != 3) stop(“missing the correct column names”)
descName <- sub(“bin”,”desc”,filename)
if(class(Inventory) == “matrix”) Inventory <- as.data.frame(Inventory)
L <- cbind(Inventory$Lat,Inventory$Lon)*(pi/180)
s <- cbind(cos(L),sin(L))
# z is the array of 3D coords of stations. All unit vecs
z <- cbind(s[,1]*s[,2],s[,1]*s[,4],s[,3])
z2 <- z/sqrt(2) # Better to mult now than after outer prod
D <- 6371*2 #Diam of Earth
n <- nrow(L)
if(n <= blockSize)stop(“BlockSize must be less than rows in Inventory”)
blocks <- n %/% blockSize
if((n %% blockSize) > 0)blocks <- blocks + 1
dex <- 1:blockSize
BM <- filebacked.big.matrix(nrow = n , ncol = n,
dimnames = list(as.list(Inventory$Id),NULL),
init = NA,
backingpath = Directory,
backingfile = filename,
descriptorfile = descName,
type = “double”)
startTime <- Sys.time()
for(i in 1:blocks){
p <- dex + (i-1)*blockSize
p <- p[p<= n]
for(j in 1:blocks){
q <- dex +(j-1)*blockSize
q <- q[q<=n]
x <- 0.5000000001-z2[p,]%*%t(z2[q,])
x <- D * asin(sqrt(x))
if(identical(p,q))diag(x)<-0
BM[p,q]<- x
}
}
print(Sys.time()-startTime)
return(BM)
}
That takes 46 seconds. orders of magintude improvement
Using bigmemory for a distance matrix
UPDATE: code is in the drop box for anybody who wants to play around. change extension from txt to R
The process of working on metadata and temperature series gives rise to several situations where I need to calculate the distance from every station to every other station. With a small number of stations this can be done easily on the fly with the result stored in a matrix. The matrix has rows and columns equal to the number of stations and it is filled by looping through the various station combinations. Of course one need only calculate the distance for every pair once. When the number of stations increases to 10s of thousands, such as with the Berkeley Earth project, there are two challenges. First is the time that the process takes and second is the memory it consumes. Handling the space problem is relatively straightforward and I decided to use bigmemory to store the data. The speed problem is an entirely different matter, but I’m making a little progress. In addition to storing the data I also want a way to quickly retrieve the neighbors of a given station and I want my solution to be relatively bullet proof and easy to understand.
To solve the problem I define two functions createDistanceMatrix() and getNeighbors(). The coed for each follows:
Lets start with createDistanceMatrix:
createDistanceMatrix <- function(Inventory,Directory = getwd(), filename= “distance.bin”){
require(bigmemory)
deg2rad <- function(deg) return(deg*pi/180)
if(!grepl(“\\.bin”,filename))stop(“please use a bin extension”)
columnsPresent <- intersect(colnames(Inventory),c(“Id”,”Lon”,”Lat”))
if(length(columnsPresent) != 3) stop(“missing the correct column names”)
descName <- sub(“bin”,”desc”,filename)
if(class(Inventory) == “matrix”) Inventory <- as.data.frame(Inventory)
The function is declared with a variable for the Inventory of stations, a Directory for where the file will be written, and a default filename. Next we indicate that the bigmemory library is required and then proceed to define a local function [deg2rad()] and do some simple input checking. I enforce the use of a specific extension (“.bin”) which I used for all my filebacked bigmatrices. Then we check the inventory to make sure that it has the columns required:columns named “Id”, “Lon” and “Lat”. In my coding Inventories are usually data.frames, but I allow for using a matrix. If you use a matrix I coerce it locally to a data.frame. Finally, I create a new file name that is critical for future “reads” of the data. In bigmemorywhen you write a file a “description file” is created to document the structure of the file. We will use this to attach the file in the future. Proceeding:
Inventory[, “sinLat”] <- sin(deg2rad(Inventory$Lat))
Inventory[, “cosLat”] <- cos(deg2rad(Inventory$Lat))
Inventory$Lon <- deg2rad(Inventory$Lon)
ER <- 6371
These lines require a bit of explanation. To calculate the distance between stations I am going to use a great circle calculation. That calculation repeatedly takes the sin() and cos() of the latitude. For speed I do that once for every station in the inventory. Also, R takes radians so I do that transformation once. ER is the earth’s radius in kilometers. Next we define some parameters for the file we want to create:
nr <- nrow(Inventory)
nc <- nrow(Inventory)
options(bigmemory.allow.dimnames=TRUE)
The matrix we create and store as a file will be square. In truth I could make the matrix one row or one column smaller, but I’ve left it like this for now. Note as well that I decide to set the options for using rownames and column names. This will allow people to use character Ids for their station Id in an inventory. That will be clearer as we progress. With those preliminaries we are ready to initialize the filebacked big matrix:
D <- filebacked.big.matrix(nrow = nr, ncol = nc,
dimnames = list(as.list(Inventory$Id),NULL),
init = NA,
backingpath = Directory,
backingfile = filename,
descriptorfile = descName,
type = “double”)
Some things to note: I set the rows and columns to be equal. I initialize the data with NA. We only write into the upper diagonal of the matrix and I want the other values to be NA. I set the dimnames so that the rownames are equal to the station Ids. They can be numbers or characters. When you seek on the file you’ll use the station ID. So, you could use characters ( “USC26541328”) for station Ids and the functions will still work. I also set a directory where the file is written, the file name and the decriptor name. Note, the decriptor name is DERIVED from the filename by simply changing the extension: “distance.bin” has a file descriptor of “distance.desc”. Type is set as “double.” Next I do a couple of things that might look confusing. Sometime a station has no Latitude or Longitude. I handle that here by allowing those stations including in the Inventory, but I detect that problem and write a negative distance in the appropriate rows and columns. Our matrix will have 3 values: distance, a negative filler, and NA for elements of the matrix that dont have to be calculated ( lower half of the diagonal ). I also determine which rows have valid Lat and Lon. That will be used as a set for looping controls. Below we set the row and column of stations that have missing Lat/Lon to -9999.
missingLonLat <- which(is.na(Inventory$Lat) | is.na(Inventory$Lon))
validLonLat <- which(!is.na(Inventory$Lat) & !is.na(Inventory$Lon))
D[missingLonLat,] <- (-9999)
D[,missingLonLat] <- (-9999)
Time to loop over the data. The approach is simple: Imagine we had a 4×4 matrix: we want to generate 1:2;1:3;1:4;2:3;2:4;3:4. Our first loop control goes over the 1-3 and the second goes over 2-4. Because some stations have missing Lat/lon we will skip over them. That is the purpose of validLonLat. It conatins those rows that are valid.
for(i in validLonLat[-length(validLonLat)]){
for(j in validLonLat[validLonLat > i] ){
D[i,j] <- acos(Inventory$sinLat[i] * Inventory$sinLat[j]
+ Inventory$cosLat[i] * Inventory$cosLat[j]
* cos(Inventory$Lon[j]-Inventory$Lon[i])) * ER
if(is.nan(D[i,j])) D[i,j] <- 0
}
}
So, we start looping over the first element in validLonLat and we loop to N-1. In the second loop, we skip lower rownumbers. The range value is calculated by using the precomputed values for sinLat() and cosLat() so that I am basically reducing the number of trig functions that have to be called by orders of magnitude. Finally, there is a nasty little warning we have to care about. Sometimes when two stations have the same location acos() will blow up. This happens when acos() is feed a number slightly greater than 1 or less than -1. It happens rarely. The result is a NaN instead of the desired result of zero. Rather than prevent the warning, I decide to allow the calculation and catch and fix the error. I suppose I could trim sinLat or cosLat and prevent it. In anycase when this finishes we have created a bigmatrix on file. Lets test it:
Inventory<-data.frame(Id=seq(1,1000),Lat=runif(1000,-90,90),Lon= runif(1000,-180,180))
Inventory$Lat[unique(sample.int(1000,20))]<-NA
Inventory$Lon[unique(sample.int(1000,20))]<-NA
startTime <- Sys.time()
BD <- createDistanceMatrix(Inventory)
print( Sys.time()-startTime)
I create a random inventory of 1000 stations. I introduce some random NAs and time the function:
Time difference of 6.371714 mins
Creating a matrix of 1 million elements takes a bit over 6 minutes. For this we are calculating roughly 500K distances. I’m sure part of the slowness of this approach is the repeated writing I am doing to the file. So, I’ll have to investigate ways to buffer this, but for now it works. I could also see ways of using apply(). Now, lets look at how we get data from this file.
test <- getNeighbors(“distance.desc”, Id = 333)
Warning message:
In getNeighbors("distance.desc", Id = 333) : coercing Id to character
To grab a stations neighbors I call “getNeighbors” and pass the filename of the descriptor file. I may change this, bt you’ll get the idea. I also pass the Id. Id is supposed to be passed as a character. If you screw up and pass an integer, I coerce it under the hood and proceed. The code for the function looks like this:
getNeighbors <- function(descriptFile,Id){
require(bigmemory)
D <- attach.big.matrix(dget(descriptFile))
if(class(Id) != “character”){
warning(“coercing Id to character”)
Id <- as.character(Id)
}
dex <- which(rownames(D) == Id)
if(length(dex) !=1) {
if(length(dex) == 0)stop(“index not found”)
if(length(dex) > 1)stop(“index found multiple times”)
}
neighbors <- c(D[1:dex,dex],D[dex,(dex+1):ncol(D)])
neighbors[is.na(neighbors)] <- 0
neighbors[neighbors < 0] <- NA
names(neighbors) <- rownames(D)
return(neighbors)
}
Pretty straightforward. First we “attach” the file to read it using the descriptor file. If “Id” is not a character I coerce it. Then I do some checking to insure that the ID can be found in the rownames of the file and to make sure it is only found once. I will probably add a check to the creating file to make sure that Ids are unique. For now I check it here, but that will change. Then I pull the neighbors for the ID. The neighbors of any given station consists of the cells in a column and cells in a row. The best way to see this is to draw a diagram using a 4×4 matrix as an example: presume you want the 3rd row: To get those values you take rows 1-3 of the 3rd column. cell 3,3 will be NA because we do not calculate the distance from a site to itself. The balance of neighbors will be in 3 row on column 4. neighbors <- c(D[1:dex,dex],D[dex,(dex+1):ncol(D)])
That line collects all the neighbors for an Id including the Id. Next, we take the cell that is NA D[dex,dex] and set it to 0 in neighbors. Finally we take the negative distances and set them to NA. I suppose I could just as easily get rid of the whole setting things to -9999 ploy and probably will in a final version.
Lets look at test:
test[330:340]
330 331 332 333 334 335 336 337 338
7722.563 NA 7564.581 0.000 16180.166 8846.114 15410.572 5890.057 17409.563
339 340
7262.202 10958.382
So, station 331 had a missing Lat/Lon and station 333 is zero km from itself.
Neat. Ok. Taking a few minutes to add some clean up. Here is the revised versions
createDistanceMatrix <- function(Inventory,Directory = getwd(), filename= “distance.bin”){
require(bigmemory)
deg2rad <- function(deg) return(deg*pi/180)
if(!grepl(“\\.bin”,filename))stop(“please use a bin extension”)
columnsPresent <- intersect(colnames(Inventory),c(“Id”,”Lon”,”Lat”))
if(length(columnsPresent) != 3) stop(“missing the correct column names”)
descName <- sub(“bin”,”desc”,filename)
if(class(Inventory) == “matrix”) Inventory <- as.data.frame(Inventory)
if(length(unique(Inventory$Id)) != length(Inventory$Id))stop(“Ids, must be unique”)
# some temporary variables to speed up processing of distance calculation
# these are done once and resused inside the loop
Inventory[, “sinLat”] <- sin(deg2rad(Inventory$Lat))
Inventory[, “cosLat”] <- cos(deg2rad(Inventory$Lat))
Inventory$Lon <- deg2rad(Inventory$Lon)
ER <- 6371
nr <- nrow(Inventory)
nc <- nrow(Inventory)
options(bigmemory.allow.dimnames=TRUE)
D <- filebacked.big.matrix(nrow = nr, ncol = nc,
dimnames = list(as.list(Inventory$Id),NULL),
init = NA,
backingpath = Directory,
backingfile = filename,
descriptorfile = descName,
type = “double”)
validLonLat <- which(!is.na(Inventory$Lat) & !is.na(Inventory$Lon))
for(i in validLonLat[-length(validLonLat)]){
for(j in validLonLat[validLonLat > i] ){
D[i,j] <- acos(Inventory$sinLat[i] * Inventory$sinLat[j]
+ Inventory$cosLat[i] * Inventory$cosLat[j]
* cos(Inventory$Lon[j]-Inventory$Lon[i])) * ER
if(is.nan(D[i,j])) D[i,j] <- 0
}
}
return(D)
}
And
getNeighbors <- function(descriptFile = “distance.desc”,Id){
require(bigmemory)
DATA <- attach.big.matrix(dget(descriptFile))
if(class(Id) != “character”){
warning(“coercing Id to character”)
Id <- as.character(Id)
}
dex <- which(rownames(DATA) == Id)
if(length(dex) == 0)stop(“index not found”)
if(length(dex) > 1)stop(“index found multiple times”)
neighbors <- c(DATA[1:dex,dex],DATA[dex,(dex+1):ncol(DATA)])
neighbors[dex] <- 0
names(neighbors) <- rownames(DATA)
return(neighbors)
}
Metadata Dubuque and UHI
I’m in the process of remaking all the metadata from scratch and looking once again at the question of UHI. There are not any global conclusions we can draw from the data yet; I’m just in the process of checking out everything that is available that could be used to illuminate the problem. The problem, as I see it, is as follows. We know from numerous studies that UHI is a real phenomena. But, it’s far from simple. First lets get down to causes. The causes of UHI according to Oke are as follows. Note 20 + years of research has no expanded this list in any significant way
In the context of global temperature we are concerned with canopy layer UHI, that is UHI below the top of buildings where surface station thermometers are. Lets go through the causes one by one.
1. Increased absorption of short wave radiation. As Oke notes the construction of buildings adds surface area to the urban landscape. Building walls absorb SW radiation and the arrangement of building can lead to multiple reflection.
2. Increased LW radiation, primarily from air pollution over the city
3. decreased LW radiation from the surface. Skyview factor is critical here and skyview is directly related to the geometry of buildings. In a flat open plane free of building the surface has a clear view of the sky and can radiate accordingly.
4. Anthropogenic heat: this is excess heat from buildings and traffic. This is should be noted changes dramatically with the type of city and with latitude. The ranges of excess watts is rather large between city types.
5. decrease evapotranspiration: This results from changing the surface of the earth.
6. decreased turbulant heat transport. Again, building geometry plays an important roll here as does the local wind.
In addition to these variable that one can control when developing and urban area there are several that cannot be controled: the wind, clouds and nearby water.
With that in mind, let’s start to look at some metadata for Dubuque and its nearby airport. First a google map view
In the following charts the city pin will be marked witha blue cross and the airport with a red cross. In terms of incoming solar we should be aware that changes to evapotranspiration can cause changes in cloud cover. Looking at Modis cloud cover, we see the count for days of cloud cover. Less clouds is more sun.
Next we want to look at transformations to the surface. In the map that follows we have marked the urban built areas as Green bits
Next we look at population. There are a few good sources here: first is a 5 minute source for 2005 and after a 2.5 minute source for the year 2000
Next, we will look at daytime LST or land surface temperature.
And next we look at Night time LSt when UHI is supposed to be the highest
In case you are wondering what that long green patch of warmth is, it’s water.
A couple things are clear. A reader named sunshine suggested that perhaps cars from the city infected the airport with UHI. Looking at the data, the differences in temperature would seem more likely to derive from fewer clouds over the city, more built surfaces, and the higher population that created those surfaces. The surfaces hold heat and they appear to retard the formation of clouds, leading to more sunshine in the city than at the airport. And, it seems the nearby water is morel likely to modulate the temperature at the airport than anything else.
Funny. First “sunshine” thought it was “depopulation”. Then he surmised it was cars. In all of this there was no attempt to to verify a thesis, just random thoughts. Sunshine, appears to have his head where there is none. In any case trying to find a cause for cooling trends or a cause for warming trends, trying to find a UHI signal, isn’t a simple task of throwing up random thoughts. We know the causes. 20 years after Oke’s essay, the causes remain the same. The weight of the causes is critical. more on that later
Cooling Stations
Over the course of the past few years I have taking a look at cooling stations a few times but never in much depth. A few other people have looked at them, but usually without much rigor. The standard approach is to find some cooling stations and then conclude that somehow global warming is thereby challenged as a theory. I supp that argument will come up in the comments, but my real goal here is to shed some light of the phenomena of stations that have long term cooling trends. The dataset I used here is the Berkeley Earth Data set version 2.0. I used Single value, with Quality control and seasonality left in the dataset.
The data selection process is pretty simple. We read the data in, window it to 1900 to 2011, and remove those stations that have no data
Data <- readAsArray(Directory = choose.dir())
Data <- windowArray(Data, start = 1900, end = 2011.99)
Data <- removeNaStations(Data)
In the next step I take notice of something that Carrick suggested over at Lucia’s and I focus on stations that have a high percentage of complete data. For this exercise I slected those records that had 99% of the data present for the time period. That’s 1332 months of data in this 112 year period (1344 months)
rsum <- rowSums(!is.na(Data))
dex<- which(rsum > 1331)
Data <- Data[dex, , ]
Next we align the station inventory with the data, do inverse weighting and calculate the least squares fit
Data <- intersectInvData(Stations,Data)
weights <- inverseDensity(Data$Inventory,Data$Array)
Temps <- solveTemperature(Data$Array,weights)
TIME <- seq(1900,2011)
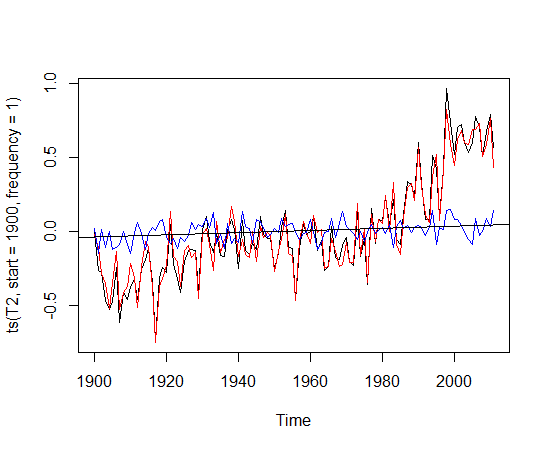
plot(ts(Temps, start =1900, frequency = 1))
abline(reg = lm(Temps~TIME))
What we see should come as no surprise. The planet is getting warmer. The slope of the line is roughly .08C/ decade. The stations have the following geographical distribution:
The sum total of all stations is 478. The result, as we expect, is similar to the results we get if we use all stations, regardless of their length. However, we can note that the sample isn’t very representative so their could be bias due to spatial sampling. The overweighting of the northern hemisphere is likely to cause a trend that is slightly higher than a what we would see if the sample included more SH stations.
The next step I will merely describe in words because I will probably change the code I used here when I decide upon a final approach. After calculating the global average of all these long stations I then created trends and confidence intervals for every station. For trends I used a Theil-Sen estimator which may be a bit different than most folks are used to. Its described here: http://en.wikipedia.org/wiki/Theil%E2%80%93Sen_estimator. The zyp package has a quick implementation that spits out confidence intervals as well. I’ll play around with some of Lucia’s approaches and maybe look at Tamino’s approach, but for now, this will do.
Summarizing the trends we see the following:
summary(DF$Slope) Min. 1st Qu. Median Mean 3rd Qu. Max. -0.014450 0.001390 0.006038 0.006151 0.010850 0.036920
Note that the simple meanof all slopes is less than the area weighted global solution. And note that some of the slopes of individual stations are negative. These are so called cooling stations: A crude map below gives you an indication of their locations. Blue is cooling, red is warming. The warming were drawn first so they tend to get covered up.
Well there you have it. Some long stations cool. This is fairly well known but a week doesnt pass on the internet without somebody asking about it or pointing to it as proof of “something.” Before we dive into that there is a bit more work to do.
In order to look a bit deeper into the “cooling stations” one beneficial cut to make through the data is to isolate those stations that are coastal versus those that are inland. That’s interesting in and of itself. The effects of geography on climate are well known. Coastal stations will have a fewer extremes. In mathematical terms the standard deviation of monthly temperatures will be lower. Defining a range for coastal stations is somewhat problematic so I did some sensitivity analysis around that looking at the standard deviation as a function of range from the ocean. More on that in the future but for now Here are the results at 50km
And now for inland stations
An interesting question this raises is could their be a second process at work with a slightly negative mean trend. The first step in looking into that will be separating those stations which have a statistically significant cooling trend from those that do not. That is, before we set out looking for a “cause” behind the cooling stations it’s probably a wise move to look at those where the cooling trend is statistically significant. Of the 478 stations, 54 show a rate of cooling that is statistically significant (95%)
With these 54 stations and the metadata we have we can start to eliminate potential causes of the cooling. One of the first thoughts that occurs to people is “de urbanization” This has been raised several times over the past 5 years I’ve spent looking at the problem. In Our AGU poster I spent a short period of time looking at “de urbanization” and frankly didn’t find much. The number of stations that had decreases in population was very small. Nevertheless, we can look at it again very quickly for these 54 stations:
First, the historgram of year 2000 population density ( people per sq km), second the histogram for 1900, and then finally the histogram for the difference between 2000 and 1900:
As the chart shows there is only one station that had a decrease in population. That station started with 16 people per sq km in 1900 and slowly over time reduced to 14 people per sq km. Lets look at some of its metadata
Slope Upper Lower Id Lat Lon Altitude Months Dst
31137 -0.0088867 -0.005273885 -0.01257143 38048 42.3978 -90.7036 325.55 1343 1212
Sdev Anthrome2000 Anthrome1900 Lcover UrbArea Slope2 Upper2
31137 2.332492 32 32 14 0.0004538235 -0.008970948 -0.005294326
Lower2 popd_1900AD popd_1910AD popd_1920AD popd_1930AD popd_1940AD popd_1950AD
31137 -0.01267606 16.7313 15.5423 15.7447 15.2845 14.9356 15.4139
popd_1960AD popd_1970AD popd_1980AD popd_1990AD popd_2000AD
31137 15.5131 15.3346 15.1406 14.1625 14.3402
The station is 1212 km away from a body of water; and we can see that its standard deviation is high. Stations near the coast have standard deviations close to 1.4. A standard deviation of 2.33, in fact, is quite high in the last quartile of all 487 stations. Other things to note. The anthrome has not changed. Anthrome 32 is residential rainfed croplands. Lcover 14 also indicates rainfed croplands ( 300 meter data). My UrbArea paramater captures the amount of “built” area within 10km, so it looks like there is some human structure in the area. In addition to this metadata, my other sources indicate an airport location nearby, so it looks like it would be an airport built in a farming community. Checking with google earth. There you go:
So, how do we explain the cooling at a station that is located at an airport? Without looking at the station history in detail, you can probably figure that the station has a station move in its history. That move may be documented or undocumented. If there is a move, that might show up in the temperature record as a discontinuity. This is a good lesson in using Berkeley Earth Surface Data. The Single value data has not been through the scalpel. The scalpel performs the analysis to find structural breaks. Series are not homogenized or adjusted, but if there is a break then a single series is replaced with two series. Let’s see what this series looks like:
Looking at the chart the step down post 1950 looks like it may be the cause of the cooling. 1950 would be the start date of the dubuque regional airport series. When I get some time I’ll load up structchange and see if it can find the discontinuity. We can continue this process for the other 53 stations, but the speculation that all the cooling is due to “de population” really doesnt have much merit. The actual number of stations that have suffered de population or de urbanization is small. The more likely explanation for the coolings comes down to undocumented or documented station changes. If you spend some time looking through all the monthly series you’ll begin to see some patterns that usually turn out to be either station moves or station splices. At some point when the “post scalpel” data is released this will be a bit easier to understand.
More Anthromes !
First off let me thank folks for all the comments and suggestions. I’m just starting to explore this data so perhaps I should explain how I go about doing this. First off, I am looking for a global bias in the record from UHI. It is well known that you can look through the data and stations and find “cases” where an urban station looks to have UHI. Doing this is easy. Pick the biggest cities you can. But, that doesnt get you an answer to the question: “what is the bias in the total record?” If you like remove those few bad apples and the answer you are left with is indistinguishable from the answer you get with those bad apples left in the record. Analytically, I prefer to take them out. However, the effect of removing them or including them is minimal. The concern is that the UHI we see in large cities ( more than 1M pop) is also present in smaller cities and perhaps even in villages.
After looking at the global averages of “urban” and non urban, the next step I like to take is to look at long series. I selected urban stations, but I did not require that the station have data during the entire period. In this test I test that decision. For this test I do the following
1. Select all stations that are “urban” in the year 2000
2. Window the data to 1900 to 2011
3. Select only those stations that have 1000 months of data. In this 112 year period they have to have 1000 of 1344 months present or about 75%
4. create 2 inventories:
A: start as urban in 1900 and end as urban in 2011: 474 stations
B: start as non urban and end as urban: 950 stations
Here are the maps for these stations: Urban designates “starts as urban”; Rural designates “starts as Rural” And dont bug me about graphics. I’m just exploring stuff here.
Next I show you a histogram of the “starting Anthrome” of the “rural” stations. Sorry no legend, I’ll explain in the text
The Anthrome key is as follows
21: Rice villages
22: Irrigated villages
23: Rainfed villages
24: Pastoral villages
31: Residential irrigated croplands
32: Residential rainfed croplands
33: Populated croplands
34: Remote croplands
41: Residential rangelands
42: Populated rangelands
43: Remote rangelands
51: Residential woodlands
52: Populated woodlands
53: Remote woodlands
54: Inhabited treeless and barren lands
61: Wild woodlands
62: Wild treeless and barren lands
What we see is that the sample is dominated by residential rainfed croplands being transformed into urban areas. There are a fair number of rice lands transformed and wooded areas transformed. This is interesting because we know from Imhoff that transforming wooded areas to urban areas yeilds the highest UHI. Why is this important? When you read a UHI study that shows a large UHI understand that the UHI is a function of the type of surrounding biome. Clearing woodland to make a city creates a high UHI– a HIGH differential from the surrounding area. creating cities in other biomes doesnt create a differential that is as high. A city in arid areas has no UHI according to Imhoff. In any case, there is more work to do here.
So, what’s the answer? Looking at long series ONLY, what happens to the warming signal when we shift land use and population from non urban( mostly croplands) to Urban?
This should look familiar. black curve is urban, red curve is “non urban” . Blue curve is the difference and I draw the slope of the difference. That slope is indistinguishable form the global answer. 0007C/year
More work I suppose
Anthromes and UHI
With BerkeleyEarth 1.6 posted to CRAN I figured it was time to do some sample programs to explain how the package worked and integrated with other packages. Also, I have some issues to check out with the metadata; and in the long run I want to reformulate my metadata package to include some new resources. There is a great project I found at http://www.worldgrids.org. When that project completes I think it will make an interesting addition to our understanding of temperature station metadata. As I travelled around the internet in search of more data I happened upon a couple of resources. First a resource that will make a great cross check on Modis Urban extent data: It’s a 300 meter resolution map of the entire globe: http://postel.mediasfrance.org/sommaire.php3?langue=English. The dataset is behind a registration wall, but getting access to it was easier than getting access to Modis. In a future post I will compare the two datasets. The second dataset I found was referenced at worldgrids: Anthromes. What is an anthrome? Human beings, like other animals, transform the landscape they inhabit, and anthromes represent the various classes of human transformed landscape. In a nutshell an Anthrome is a function of population, technology, affluence and the natural landscape. Croplands are an example. So are cities. This method of classifying the lanscape was interesting to me because of the difficulty researchers have had in finding a discernable UHI signal in global temperature records. I won’t recount the list of failed attempts. In simple terms most researchers attack the problem by identifying “rural” sites and “urban” sites and then looking for differences in trends between the two. That is relatively straightfoward. If “urban” sites are infected with UHI, then we should be able to see that bias by differencing rural sites and urban sites. That approach has several complications. First and foremost is the problem of how one classifies a site as Urban or Rural. Underneath that is the assumption that rural sites are not biased by land use changes themselves. Anthromes, it seemed to me, might provide a way out of this issue as sites are classified by basically two dimensions– a land use dimension and a population dimension. The data is open and freely available and can be read into R rather simply after downloading.
Anthrome data is delivered in 5 minute resolution or roughly a 10KM cell –at the equator. In addition, there are historical estimates for 1900, 1800 and 1700. The work depends upon the HYDE 3.1 dataset and Landscan population. The Anthromes are shown below in the legend
I started with a rather simple test. Let’s go the code
require(BerkeleyEarth)
Stations <- readSiteSummary(Directory = choose.dir())
Anthromes1900 <- file.choose()
Anthromes2000 <- file.choose()
Ant1900 <- raster(Anthromes1900)
Ant2000 <- raster(Anthromes2000)
lonlat <- cbind(Stations$Lon, Stations$Lat)
Stations <- cbind(Stations, Anthrome1900 = extract(Ant1900,y = lonlat),
Anthrome2000 = extract(Ant2000,y= lonlat))
Urban <- Stations[which(Stations$Anthrome2000 == 12 | Stations$Anthrome2000 ==11),]
Pretty simple. I use the BerkeleyEarth function to read in the stations. Then I read in two rasters: one containing Anthromes for 1900 and the other for the year 2000. Then I extract the values for each station given its Longitude and Latitude. Then I select only those stations which were Urban or mixed settlements in the year 2000. That reduces the 36,000 Berkeley Earth stations to around 8000. Their geographic distribution looks like this.
Next, we read in the data, window it to 1900 to 2011 and then we use “intersectInvData()” to make sure that the stations in the inventory match those in the data. After that we calculate weights and then “solve” a least squares problem.
Data <- readAsArray(Directory = choose.dir())
Data <- windowArray(Data, start = 1900, end = 2011.99)
Data <- intersectInvData(Urban, Data)
weights <- inverseDensity(Data$Inventory,Data$Array)
Temps <- solveTemperature(Data$Array,weights)
That’s it! The variable Temps now holds the temperature anomalies for all the urban anthromes. Next, I decided to segregate the Urban Anthromes into 2 classes: those that were already urban in 1900 and those that were not. So, we have two classes. In one class the stations were urban in 1900 and remained urban in 2000. In the other class, they were non urban in 1900 and become urban in 2000. I solve the least squares equation for both sub classes and difference them.Think about what you expect the answer to be. In short, you will have two curves. The black curve is the temperature anomaly of all the urban stations that remained urban. The red line is the anomaly of the non urban stations that became urban.
The black line represents those stations that began as Urban and ended as Urban. The red line is stations that began as non urban and became urban. There is a tiny positive trend (.0007) in this difference. What does that mean? You tell me. Did you expect stations that went from non urban to urban to show more warming? I did. Can this negative result be explained by appealing to a variety of suppositions? Sure. As always with these things more research required.
BerkeleyEarth Version 1.6
Version 1.6 has just been submitted to CRAN and I will post the source here in the dropbox. Version 1.6 will probably be stable for a long time unless I find a bug. The main additions I made were adding a few functions to make some things easier, and I converted two files–flags and sources– to file.backed.big.matrixes. Thus the read() routines for those files have changed the input parameters. The flags.txt and the sources.txt files are so large that they cause even my 4GB windows system to choke and slow down. The functions now work to create a *bin file when they are first called. After the first call, access to them is immediate. In addition I added a function for creating *bin files for all versions of data.txt you may have on your system. makeBinFiles() is called automagically after you download all the files using downloadBerkeley(). Or, you can call it separately from your main working directory. Converting the files takes about 10 minutes per file but it worth the time to do it once. After that, the file is attached instantly.
With 1.6 installed let’s do a short example to illustrate something about the Berkeley datasets. I’ve installed the package 1.6 and created a working directory called BestDownloadTest. I make that my working directory and I run downloadBerkeley(). After that function completes I have a workspace that looks like this:
I’ll run getFileInformation() on the TAVG directory. That function will create separate readmes for the all the files and collect some information. Next, we can select a directory to work with, on windows I just use choose.dir() and point at a directory like the “Single-Value” folder. I can do this like so Data <- readBerkeleyData(Directory=choose.dir()) and that will instantly attach “data.bin” to the variable Data.
Then we can get some simple statistics on the file length(unique(Data[,”Id”])) gives us the number of unique station Ids: 36853. The range of dates min(Data[,”Date”]) 1701.042 and the then the max(Data[,”Date”]) 2011.875. And then I can do histograms of the dates and the temperatures